| CAT No | Product | Size | Price |
|---|---|---|---|
| PB20.11-01 | qPCRBIO SyGreen® Mix Lo-ROX | 100 x 20 μL Reactions | Contact us |
| PB20.11-05 | qPCRBIO SyGreen® Mix Lo-ROX | 500 x 20 μL Reactions | Contact us |
| PB20.11-06 | qPCRBIO SyGreen® Mix Lo-ROX | 500 x 20 μL Reactions (1 x 5 mL) | Contact us |
| PB20.11-20 | qPCRBIO SyGreen® Mix Lo-ROX | 2000 x 20 μL Reactions | Contact us |
| PB20.11-50 | qPCRBIO SyGreen® Mix Lo-ROX | 5000 x 20 μL Reactions (1 x 50 mL) | Contact us |
| PB20.11-51 | qPCRBIO SyGreen® Mix Lo-ROX | 5000 x 20 μL Reactions (50 x 1 mL) | Contact us |
| PB20.12-01 | qPCRBIO SyGreen® Mix Hi-ROX | 100 x 20 μL Reactions | Contact us |
| PB20.12-05 | qPCRBIO SyGreen® Mix Hi-ROX | 500 x 20 μL Reactions | Contact us |
| PB20.12-06 | qPCRBIO SyGreen® Mix Hi-ROX | 500 x 20 μL Reactions (1 x 5 mL) | Contact us |
| PB20.12-20 | qPCRBIO SyGreen® Mix Hi-ROX | 2000 x 20 μL Reactions | Contact us |
| PB20.12-50 | qPCRBIO SyGreen® Mix Hi-ROX | 5000 x 20 μL Reactions (1 x 50 mL) | Contact us |
| PB20.12-51 | qPCRBIO SyGreen® Mix Hi-ROX | 5000 x 20 μL Reactions (50 x 1 mL) | Contact us |
| PB20.13-01 | qPCRBIO SyGreen® Mix with Fluorescein | 100 x 20 μL Reactions | Contact us |
| PB20.13-05 | qPCRBIO SyGreen® Mix with Fluorescein | 500 x 20 μL Reactions | Contact us |
| PB20.13-06 | qPCRBIO SyGreen® Mix with Fluorescein | 500 x 20 μL Reactions (1 x 5 mL) | Contact us |
| PB20.13-20 | qPCRBIO SyGreen® Mix with Fluorescein | 2000 x 20 μL Reactions | Contact us |
| PB20.14-01 | qPCRBIO SyGreen® Mix Separate-ROX | 100 x 20 μL Reactions | Contact us |
| PB20.14-05 | qPCRBIO SyGreen® Mix Separate-ROX | 500 x 20 μL Reactions | Contact us |
| PB20.14-06 | qPCRBIO SyGreen® Mix Separate-ROX | 500 x 20 μL Reactions (1 x 5 mL) | Contact us |
| PB20.14-20 | qPCRBIO SyGreen® Mix Separate-ROX | 2000 x 20 μL Reactions | Contact us |
| PB20.14-50 | qPCRBIO SyGreen® Mix Separate-ROX | 5000 x 20 μL Reactions (1 x 50 mL) | Contact us |
| PB20.14-51 | qPCRBIO SyGreen® Mix Separate-ROX | 5000 x 20 μL Reactions (50 x 1 mL) | Contact us |
To see your pricing, please log in or register with your quote reference.
Additional information
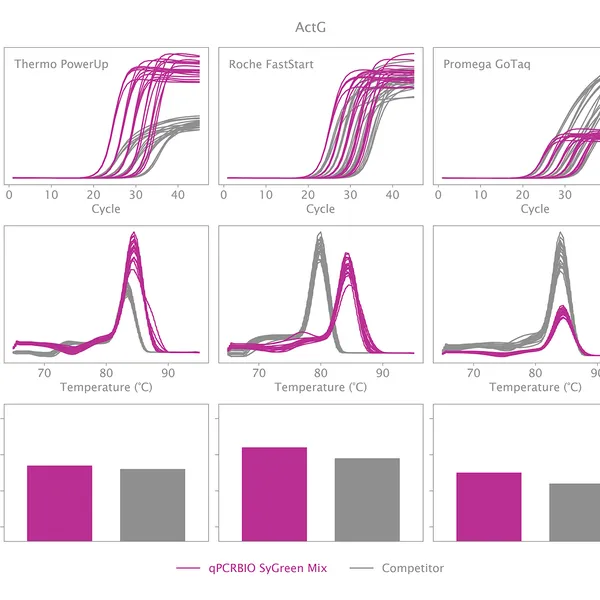
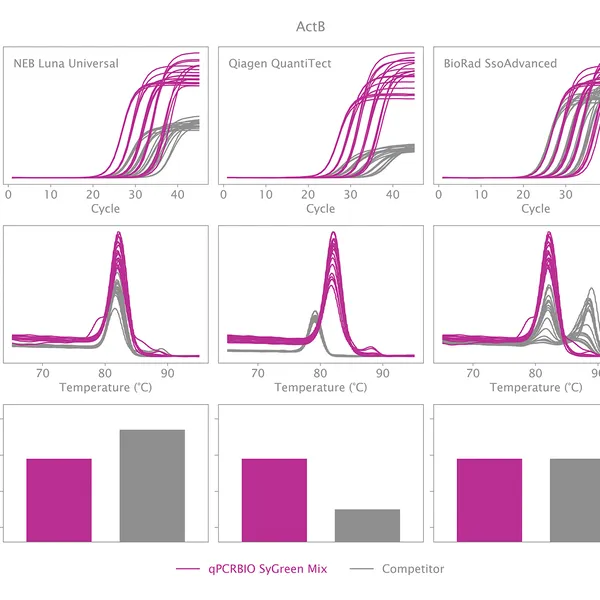
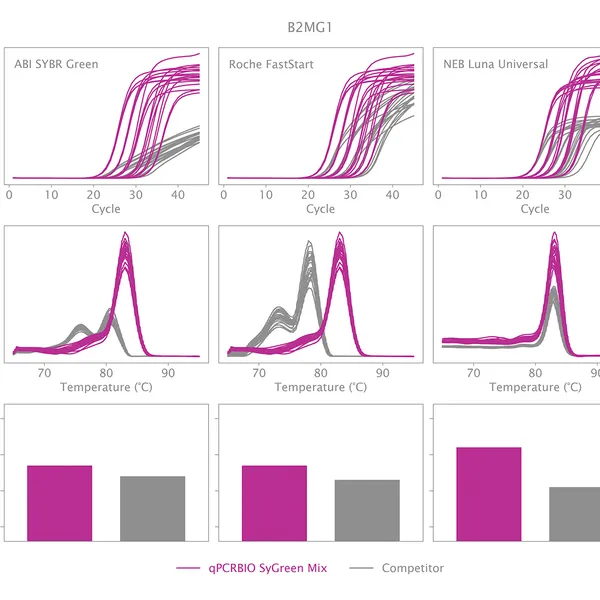
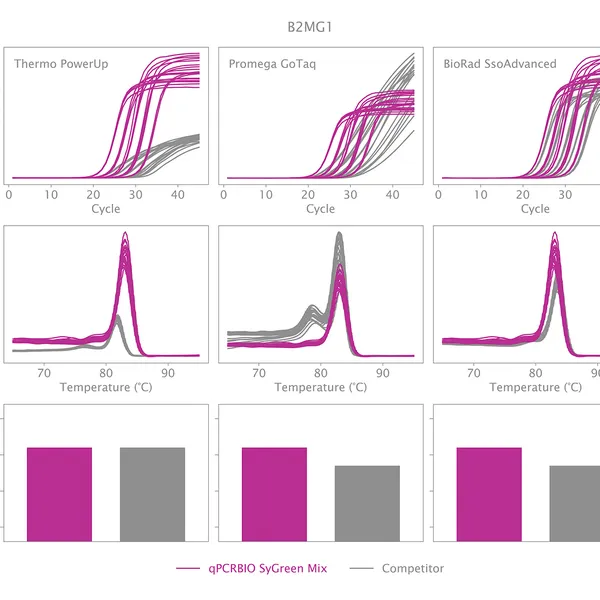
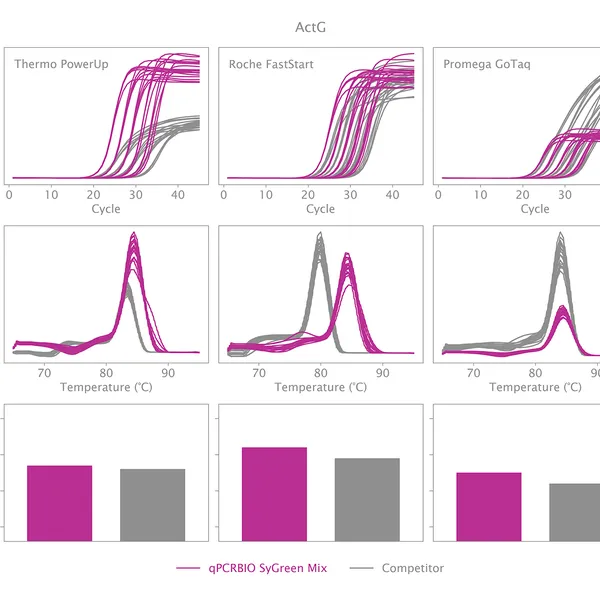
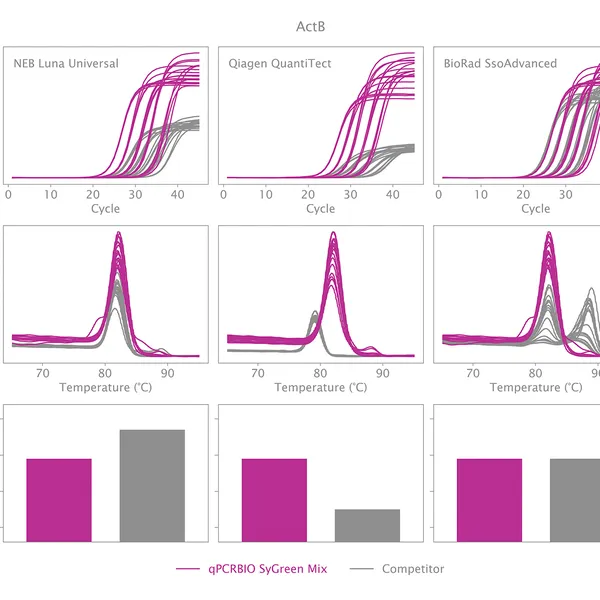
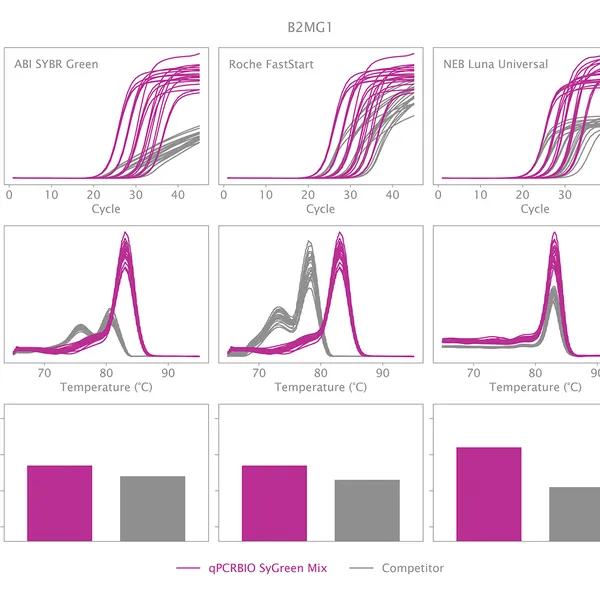
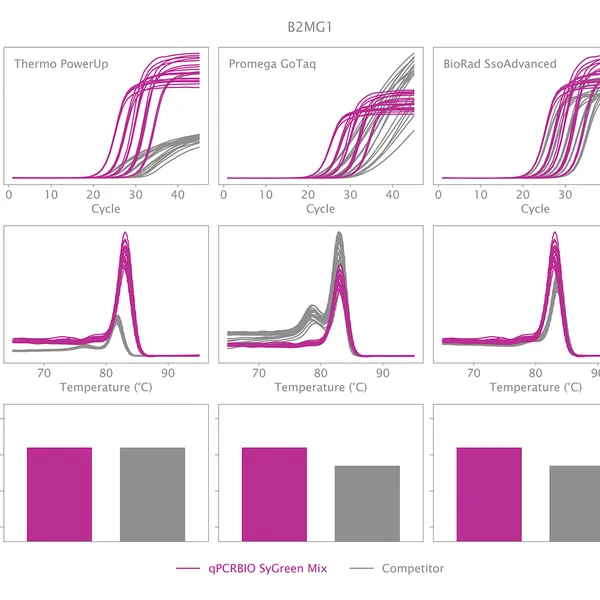
qPCRBIO SyGreen® Mix is designed for fast, highly sensitive, and reproducible dye-based qPCR with minimal or no optimization. Hot start technology via antibody prevents the formation of primer dimers and non-specific products, resulting in significantly improved reaction sensitivity and specificity.
Our 2x mix uses a proprietary DNA intercalating dye, non-inhibitory to PCR, unlike many common fluorescent dyes. The improvements that increase the sensitivity and homogeneity of qPCRBIO SyGreen® Mix in standard cycling conditions allow for leading performance in rapid and ultra-fast cycling conditions as well.
Use our qPCR Selection Tool to find out which ROX variant is compatible with your equipment.
How does qPCRBIO SyGreen® work?
These intercalating dyes contain aromatic and flat groups allowing the process of intercalation of the molecules into the minor groove of double-stranded DNA spanning about 3.5-4 nucleotides. The presence of positive charges on the dyes stabilizes them, allowing coordination with the negatively charged DNA backbone.
Due to this DNA-binding mechanism, they cannot bind to single-stranded nucleic acids unless secondary structures are present. On the other hand, fluorescence is increased up to 1,000-fold when intercalated with dsDNA.
When a dye intercalates between DNA base pairs, it emits fluorescence proportional to the number of bound dyes (and thus the amount of DNA in the sample). This allows the visualization of nucleic acids on a gel or in a fluorimeter in real-time qPCR devices. In the latter case, the number of dye molecules intercalating in the sample increases with qPCR, leading to a corresponding increase in fluorescence with each cycle. At the beginning of the reaction, the free dye emits a baseline level of fluorescence. As the reaction progresses, the fluorescence intensity increases exponentially, before stabilizing in later cycles, leading to the classic S-shaped amplification curve.
Advantages and limitations of dye-based qPCR
Advantages:
- Diverse Target Detection: By combining a SyGreen dye or SYBR Green type with primers, studies can detect any molecular target. This flexibility allows for efficient and straightforward singleplex qPCR design.
- Reversible Fluorescence Signal Accumulation: Using DNA intercalating dyes facilitates the reversible accumulation of fluorescence signals. Since strong fluorescence only arises from intercalated dye, DNA denaturation results in a reduction of fluorescence signal. This characteristic allows specificity experiments through melting curve analysis, enabling rapid evaluation of the specificity of qPCR.
Limitations:
- Non-specific Signal Generation: Intercalating dyes can produce non-specific fluorescence signals as they bind to any dsDNA molecule in the tube. This can result from non-specific primer binding, the presence of similar sequences within the sample, primer-dimer formation, or potential stem-loop primer structures. To verify results, additional analyses such as melting curve analysis, gel electrophoresis, or Sanger sequencing are recommended.
- Single Target Assay: Dye-based qPCR only allows the analysis of one target per reaction. To quantify multiple targets in one sample, the corresponding number of assays must be set up.
Melting Curve Analysis
Melting curve analysis serves to determine the melting point of the qPCR product, providing valuable information on product specificity. This analysis involves an additional heat cycle step at the end of the qPCR cycling program. In this step, reaction samples are gradually denatured by increasing incubation temperature while collecting fluorescence data at each temperature interval. Plotting fluorescence intensity data against temperature results in a melting curve. In a reaction containing specific products, fluorescence intensity remains stable until the temperature approaches the target's melting point, at which point it rapidly drops to baseline levels. The temperature at which fluorescence intensity drops to 50% of the maximum is considered the melting point or Tm of that molecule. The specificity of the reaction can be simply assessed by plotting the first derivative of fluorescence intensity against temperature change.
For more detailed information on dye-based qPCR or qPCR in general, please refer to our technical guide.
Note:
SYBR Green is a registered trademark of Molecular Probes Inc. and owned by Thermo Fischer Scientific. PCR Biosystems Ltd. is not affiliated with any of these entities.








Read more